NP-Sticky: Model-Guided Ligation Calculator
Sticky End Finder
The sticky end finder is designed to find the optimal sticky ends from existing sequences. Sticky ends are ranked based on the discrimination between correct and misannealed free energies of ligation. A higher score indicates fewer misannealed structures.
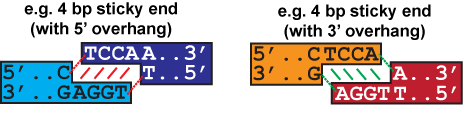
In both examples above, the sticky end overhang is TCCA. The sticky end finder is most accurate between sticky ends with the same number of overhang bases. One application of the sticky end finder is to identify or create the best cloning sites in a display vector.
Please input DNA sequence (5'→3' upper strand) into the box below.
 This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.The Python source code is available here: se_finder.cgi
Last modified by Daphne Ng on Jan 14th, 2014